

| Statistics Toolbox |   |
N-way Analysis of Variance (ANOVA)
Syntax
p = anovan(X,group) p = anovan(X,group,'model') p = anovan(X,group,'model',sstype) p = anovan(X,group,'model',sstype,gnames) p = anovan(X,group,'model',sstype,gnames,'displayopt') [p,table] = anovan(...) [p,table,stats] = anovan(...) [p,table,stats,terms] = anovan(...)
Description
p = anovan(X,group)
performs a balanced or unbalanced multi-way ANOVA for comparing the means of the observations in vector X with respect to N different factors. The factors and factor levels of the observations in X are assigned by the cell array group. Each of the N cells in group contains a list of factor levels identifying the observations in X with respect to one of the N factors. The list within each cell can be a vector, character array, or cell array of strings, and must have the same number of elements as X.
As an example, consider the X and group inputs below.
X = [x1 x2 x3 x4 x5 x6 x7 x8]; group = {[1 2 1 2 1 2 1 2];... ['hi';'hi';'lo';'lo';'hi';'hi';'lo';'lo'];... {'may' 'may' 'may' 'may' 'june' 'june' 'june' 'june'}} ;
In this case, anovan(X,group) is a three-way ANOVA with two levels of each factor. Every observation in X is identified by a combination of factor levels in group. If the factors are A, B, and C, then observation x1 is associated with:
Similarly, observation x6 is associated with:
Output vector p contains p-values for the null hypotheses on the N main effects. Element p(1) contains the p-value for the null hypotheses, H0A, that samples at all levels of factor A are drawn from the same population, element p(2) contains the p-value for the null hypotheses, H0B, that samples at all levels of factor B are drawn from the same population, and so on.
If any p-value is near zero, this casts doubt on the associated null hypothesis. For example, a sufficiently small p-value for H0A suggests that at least one A-sample mean is significantly different that the other A-sample means; i.e., there is a main effect due to factor A. The choice of a limit for the p-value to determine whether a result is "statistically significant" is left to the researcher. It is common to declare a result significant if the p-value is less than 0.05 or 0.01.
anovan also displays a figure showing the standard ANOVA table, which by default divides the variability of the data in X into:
The ANOVA table has six columns:
p = anovan(X,group, performs the ANOVA using the model specified by 'model')
'model', where 'model' can be 'linear', 'interaction', 'full', or an integer or vector. The default 'linear' model computes only the p-values for the null hypotheses on the N main effects. The 'interaction' model computes the p-values for null hypotheses on the N main effects and the 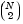 two-factor interactions. The
two-factor interactions. The 'full' model computes the p-values for null hypotheses on the N main effects and interactions at all levels.
For an integer value of 'model', k (k 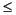
N), anovan computes all interaction levels through the kth level. The values k=1 and k=2 are equivalent to the 'linear' and 'interaction' specifications, respectively, while the value k=N is equivalent to the 'full' specification.
For more precise control over the main and interaction terms that anovan computes, 'model' can specify a vector containing one element for each main or interaction term to include in the ANOVA model. Each vector element encodes the corresponding ANOVA term as the decimal equivalent of an N-bit number, where N is the number of factors. The table below illustrates the coding for a 3-factor ANOVA.
| Corresponding ANOVA Terms |
||
| Main term A |
||
| Main term B |
||
| Main term C |
||
| Interaction term AB |
||
| Interaction term BC |
||
| Interaction term AC |
||
| Interaction term ABC |
For example, if 'model' is the vector [2 4 6], then output vector p contains the p-values for the null hypotheses on the main effects B and C and the interaction effect BC, in that order. A simple way to generate the 'model' vector is to modify the terms output, which codes the terms in the current model using the format described above. If anovan returned [2 4 6] for terms, for example, and there was no significant result for interaction BC, you could recompute the ANOVA on just the main effects B and C by specifying [2 4] for 'model'.
p = anovan(X,group, computes the ANOVA using the type of sum-of-squares specified by 'model',sstype)
sstype, which can be 1, 2, or 3 to designate Type 1, Type 2, or Type 3 sum-of-squares, respectively. The default is 3. The value of sstype only influences computations on unbalanced data.
The sum of squares for any term is determined by comparing two models. The Type 1 sum of squares for a term is the reduction in residual sum of squares obtained by adding that term to a fit that already includes the terms listed before it. The Type 2 sum of squares is the reduction in residual sum of squares obtained by adding that term to a model consisting of all other terms that do not contain the term in question. The Type 3 sum of squares is the reduction in residual sum of squares obtained by adding that term to a model containing all other terms, but with their effects constrained to obey the usual "sigma restrictions" that make models estimable.
Suppose we are fitting a model with two factors and their interaction, and that the terms appear in the order A, B, AB. Let R(·) represent the residual sum of squares for a model, so for example R(A,B,AB) is the residual sum of squares fitting the whole model, R(A) is the residual sum of squares fitting just the main effect of A, and R(1) is the residual sum of squares fitting just the mean. The three types of sums of squares are as follows:
| Term |
Type 1 SS |
Type 2 SS |
Type 3 SS |
| A |
R(1)-R(A) |
R(B)-R(A,B) |
R(B,AB)-R(A,B,AB) |
| B |
R(A)-R(A,B) |
R(A)-R(A,B) |
R(A,AB)-R(A,B,AB) |
| AB |
R(A,B)-R(A,B,AB) |
R(A,B)-R(A,B,AB) |
R(A,B)-R(A,B,AB) |
The models for Type 3 sum of squares have sigma restrictions imposed. This means, for example, that in fitting R(B,AB), the array of AB effects is constrained to sum to 0 over A for each value of B, and over B for each value of A.
p = anovan(X,group, uses the string values in character array 'model',sstype,gnames)
gnames to label the N experimental factors in the ANOVA table. The array can be a string matrix with one row per observation, or a cell array of strings with one element per observation. When gnames is not specified, the default labels 'X1', 'X2', 'X3', ..., 'XN' are used.
p = anovan(X,group, enables the ANOVA table display when ''model',sstype,gnames,'displayopt')
displayopt' is 'on' (default) and suppresses the display when 'displayopt' is 'off'.
[p,table] = anovan(...)
returns the ANOVA table (including factor labels) in cell array table. (Copy a text version of the ANOVA table to the clipboard by using the Copy Text item on the Edit menu.)
[p,table,stats] = anovan(...)
returns a stats structure that you can use to perform a follow-up multiple comparison test.
The anovan test evaluates the hypothesis that the different levels of a factor (or more generally, a term) have the same effect, against the alternative that they do not all have the same effect. Sometimes it is preferable to perform a test to determine which pairs of levels are significantly different, and which are not. Use the multcompare function to perform such tests by supplying the stats structure as input.
[p,table,stats,terms] = anovan(...)
returns the main and interaction terms used in the ANOVA computations. The terms are encoded in output vector terms using the same format described above for input 'model'. When 'model' itself is specified in this vector format, the vector returned in terms is identical.
Examples
In the previous section we used anova2 to analyze the effects of two factors on a response in a balanced design. For a design that is not balanced, we can use anovan instead.
The dataset carbig contains a number of measurements on 406 cars. Let's study how the mileage depends on where and when the cars were made.
The p-value for the interaction term is not small, indicating little evidence that the effect of the car's year or manufacture (when) depends on where the car was made (org). The linear effects of those two factors, though, are significant.
Reference
[1] Hogg, R. V. and J. Ledolter. Engineering Statistics. MacMillan Publishing Company, 1987.
See Also
anova1, anova2, multcompare
 | anova2 | aoctool |  |