

| System Identification Toolbox |   |
Compare measured outputs with model outputs.
Syntax
compare(data,m); compare(data,m,k,sampnr,init) compare(data,m1,m2,...,mN,Yplots) compare(data,m1,'PlotStyle1',...,mN,'PlotStyleN',k,sampnr,init) [yh,fit] = compare(data,m1,'PlotStyle1',...,mN,'PlotStyleN',k,sampnr,init)
Description
data is the output-input data in the usual iddata object format.
compare computes the output yh that results when the model m is simulated with the input u. The result is plotted together with the corresponding measured output y. The percentage of the ouput variation that is explained by the model
is also computed and displayed. For multi-output systems this is done separately for each output.
When the argument k is specified, the k-step ahead prediction of y according to the model m are computed instead of the simulated output. In the calculation of 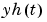 , the model can use outputs up to time
, the model can use outputs up to time 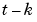 :
: 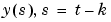 ,
, 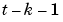 ,
, 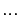 (and inputs up to the current time t). The default value of
(and inputs up to the current time t). The default value of k is inf, which gives a pure simulation from the input only.
A last argument Yplots may be given as a cell array of strings. Only the outputs with OutputName in this array will be plotted, while all are used for the necessary computations. If Yplots is not specified, all outputs will be plotted.
The argument sampnr indicates that only the sample numbers in this row vector are plotted and used for the calculation of the fit. The whole data record is used for the simulation/prediction, though.
The argument init determines how to handle initial conditions in the models:
init = 'e' (for 'estimate') estimates the initial conditions for best fit.
init = 'm' (for 'model') used the model's internally stored initial state.
init = 'z' (for 'zero') uses zero initial conditions.
init = x0, where x0 is a column vector of the same size as the state vector of the models, uses x0 as the initial state.
When several models are specified, as in compare(data,m1,m2,...,mN), the plots show responses and fits for all models. In that case data should contain all inputs and outputs that are required for the different models. However, some models may very well correspond to subselections of channels and may not need all channels in data. In that case the proper handling of signals is based on the InputNames and OutputNames of data and the models.
With compare(data,m1,'PlotStyle1',...mN,'PlotStyle2') the color, linestyle, and/or marker can be specified for the curves associated with the different models. The markers are the same as for the regular plot command. For example,
If data contains several experiments, separate plots are given for the different experiments. In this case sampnr, if specified, must be a cell array with as many entries as there are experiments.
Arguments
When output arguments [yh,fit] = compare(data,m1,..,mN) are specified, no plots are produced.
yh is a cell array of length equal to the number of models. Each cell contains the corresponding model output as an iddata object.
fit is in the general case a 3-D array with fit(kexp,kmod,ky) containing the fit (computed as above) for output ky, model kmod, and experiment kexp.
Examples
Split the data record into two parts. Use the first one for estimating a model and the second one to check the model's ability to predict six steps ahead:
See Also
 | bode | covf |  |