

| System Identification Toolbox |   |
Estimate/compute/display impulse response.
Syntax
impulse(m) impulse(data) impulse(data,'sd',sd,'pw',na,Time) impulse(m,'sd',sd,Time) impulse(m1,m2,...,dat1, ...,mN,Time,'sd',sd) impulse(m1,'PlotStyle1',m2,'PlotStyle2',...,dat1,'PlotStylek',..., mN,'PlotStyleN',Time,'sd',sd) [y,t,ysd] = impulse(m) mod = impulse(data)
Description
impulse can be applied both to idmodels and to iddata sets, as well as to any mixture.
For a discrete time idmodel m, the impulse response y and, when required, its estimated standard deviation ysd, is computed using sim. When called with output arguments, y, ysd and the time vector t are returned. When impulse is called without output arguments, a plot of the impulse response is shown. If sd is given a value larger than zero, a confidence region around zero is drawn. It corresponds to the confidence of sd standard deviations. In the plots, the impulse is inversely scaled with the sampling interval, so that it has the same energy regardless of the sampling.
Adding an argument 'fill' among the input arguments gives an uncertainty region marked by a filled area, rather than by dash-dotted lines.
The start time T1 and the end time T2 can be specified by Time= [T1 T2]. If T1 is not given, it is set to -T2/4. The negative time lags (the impulse is always assumed to occur at time 0) show possible feedback effects in the data, when the impulse is estimated directly from data. If Time is not specified, a default value is used.
For an iddata set data, impulse(data) estimates a high order, noncausal FIR model after first having prefiltered the data so that the input is "as white as possible." The impulse response of this FIR model and, when asked for, its confidence region is then plotted. When called with an output argument, impulse, in the iddata case, returns this FIR model, stored as an idarx model.The order of the prewhitening filter can be specified as na. The default value is na = 10.
Any number and any mixture of models and data sets can be used as input arguments. The responses are plotted with each input/output channel (as defined by the models' and data sets' InputName and OutputName) as a separate plot. Colors, linestyles, and marks can be defined by PlotStyle values. These are the same as for the regular plot command, like
The noise input channels in m are treated as follows: Consider a model m with both measured input channels u (nu channels) and noise channels e (ny channels) with covariance matrix 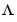
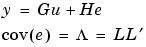
where L is a lower triangular matrix. Note that m.NoiseVariance = 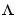 . The model can also be described with unit variance, normalized noise source v:
. The model can also be described with unit variance, normalized noise source v:
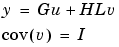
impulse(m) plots the impulse response of the transfer function G.
impulse(m('n')) plots the impulse response of the transfer function H. (ny inputs and ny outputs).The input channels have names e@yname, where yname is the name of the corresponding output.
m is a time series, that is nu = 0, impulse(m) plots the impulse response of the transfer function H.
impulse(noisecnv(m)) plots the impulse response of the transfer function [G H] (nu+ny inputs and ny outputs). The noise input channels have names e@yname, where yname is the name of the corresponding output.
impulse(noisecnv(m,'norm')) plots the impulse response of the transfer function [G HL] (nu+ny inputs and ny outputs. The noise input channels have names v@yname, where yname is the name of the corresponding output.
Arguments
If impulse is called with a single idmodel m, the output argument y is a 3-D array of dimension Nt-by-ny-by-nu. Here Nt is the length of the time vector t, ny is the number of output channels and nu is the number of input channels. Thus y(:,ky,ku) is the response in output ky to an impulse in the ku-th input channel.
ysd has the same dimensions as y and contains the standard deviations of y.
If impulse is called with an output argument and a single data set in the input arguments, the output is returned as an idarx model mod containing the high order FIR model, and its uncertainty. By calling impulse with mod, the responses can be displayed and returned without having to redo the estimation.
Example
impulse(data,'sd',3) estimates and plots the impulse response
See Also
 | idss | init |  |