

| Statistics Toolbox |   |
Pairwise distance between observations
Syntax
Description
Y = pdist(X)
computes the Euclidean distance between pairs of objects in m-by-n matrix X, which is treated as m vectors of size n. For a dataset made up of m objects, there are 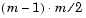 pairs.
pairs.
The output, Y, is a vector of length 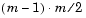 , containing the distance information. The distances are arranged in the order (1,2), (1,3), ..., (1,m), (2,3), ..., (2,m), ..., ..., (m-1,m).
, containing the distance information. The distances are arranged in the order (1,2), (1,3), ..., (1,m), (2,3), ..., (2,m), ..., ..., (m-1,m). Y is also commonly known as a similarity matrix or dissimilarity matrix.
To save space and computation time, Y is formatted as a vector. However, you can convert this vector into a square matrix using the squareform function so that element i,j in the matrix, where 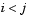 , corresponds to the distance between objects i and j in the original dataset.
, corresponds to the distance between objects i and j in the original dataset.
Y = pdist(X,'metric')
X, using the method specified by 'metric', where 'metric' can be any of the following character strings that identify ways to compute the distance.
Y = pdist(X,distfun,p1,p2,...)
accepts a function handle to a distance function of the form
taking as arguments two q-by-n matrices XI and XJ each of which contains rows of X, plus zero or more additional arguments, and returning a q-by-1 vector of distances d, whose kth element is the distance between the observations XI(k,:) and XJ(k,:). The arguments p1,p2,... are passed directly to the function distfun.
Y = pdist(X,'minkowski',p)
computes the distance between objects in the data matrix, X, using the Minkowski metric. p is the exponent used in the Minkowski computation which, by default, is 2.
Mathematical Definitions of Methods
Given an m-by-n data matrix X, which is treated as m (1-by-n) row vectors x1, x2, ..., xm, the various distances between the vector xr and xs are defined as follows:
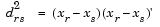
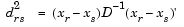
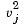 , which denotes the variance of the variable Xj over the m objects.
, which denotes the variance of the variable Xj over the m objects.
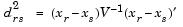
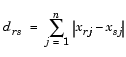
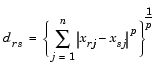
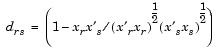
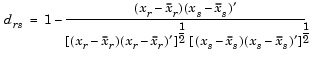
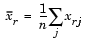 and
and 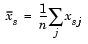
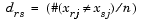
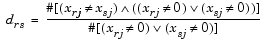
Examples
X = [1 2; 1 3; 2 2; 3 1] X = 1 2 1 3 2 2 3 1 Y = pdist(X,'mahal') Y = 2.3452 2.0000 2.3452 1.2247 2.4495 1.2247 Y = pdist(X) Y = 1.0000 1.0000 2.2361 1.4142 2.8284 1.4142 squareform(Y) ans = 0 1.0000 1.0000 2.2361 1.0000 0 1.4142 2.8284 1.0000 1.4142 0 1.4142 2.2361 2.8284 1.4142 0
See Also
cluster, clusterdata, cmdscale, cophenet, dendrogram, inconsistent, linkage, silhouette, squareform
 | perms |  |